CUTANA™ GST-MeCP2 for meCUT&RUN
Product Description
CUTANA™ GST-MeCP2 for meCUT&RUN is a GST-tagged MeCP2 methyl binding domain that is validated to enrich regions of chromatin with symmetrically methylated CpGs in CUTANA™ meCUT&RUN, a modified CUT&RUN workflow that enables streamlined, high-resolution mapping of DNA methylation. meCUT&RUN can be followed by a traditional library prep method, such as the CUTANA™ CUT&RUN Library Prep Kit (14-1001-EPC/ 14-1002-EPC), to provide ~150 bp resolution profiles of DNA methylation enrichment, or by a cytosine conversion strategy such as Enzymatic Methyl-seq (NEB® EM-seq™, preferred) or bisulfite sequencing to provide base-pair resolution of 5-methylcytosine (5mC).
Validation Data
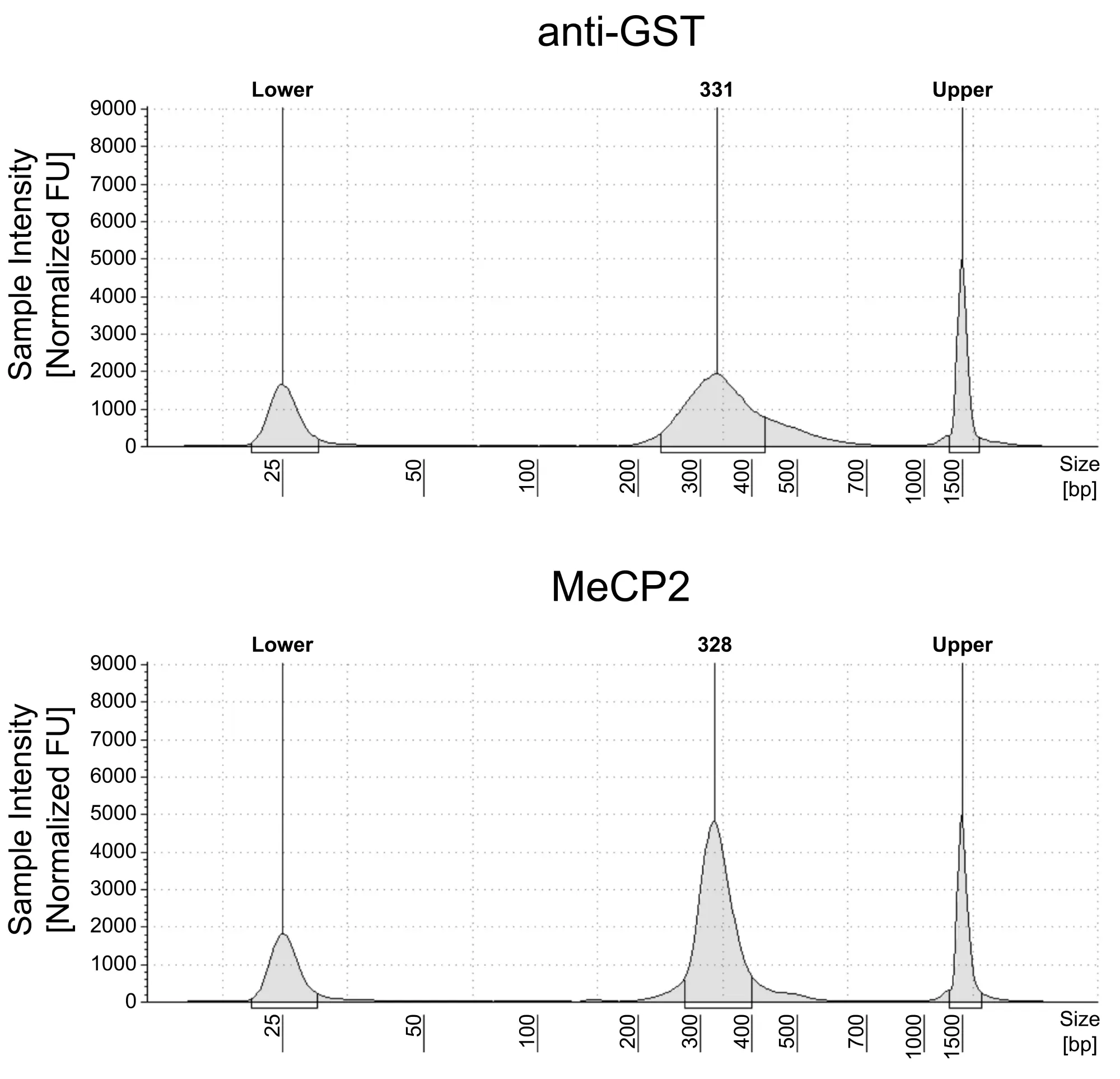
Figure 1: meCUT&RUN DNA fragment size distribution analysis
meCUT&RUN was performed as described in Figure 3. Library DNA was analyzed by Agilent TapeStation®. This analysis confirmed that mononucleosomes were predominantly enriched in meCUT&RUN (~300 bp peaks represent 150 bp nucleosomes + sequencing adapters).
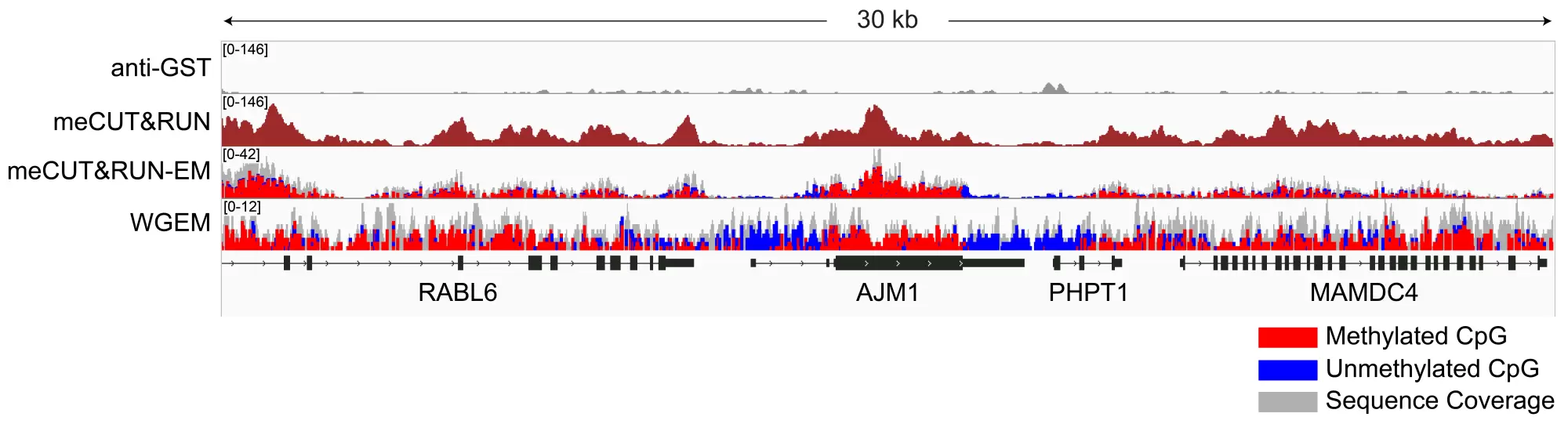
Figure 2: Gene browser tracks
meCUT&RUN was performed as described in Figure 3. A 30 kb window at the AJM1 gene is shown for anti-GST antibody and meCUT&RUN. Tracks are also shown with representative data for meCUT&RUN followed by EM-seq (meCUT&RUN-EM) and whole genome EM-seq (WGEM), using the New England Biolabs NEBNext® Enzymatic Methyl-seq v2 Kit (NEB E8015). The meCUT&RUN kit produced the expected genomic distribution, showing enrichment of methylated DNA that approximates the methylated CpG pattern observed in WGEM. Images were generated using the Integrative Genomics Viewer (IGV, Broad Institute).
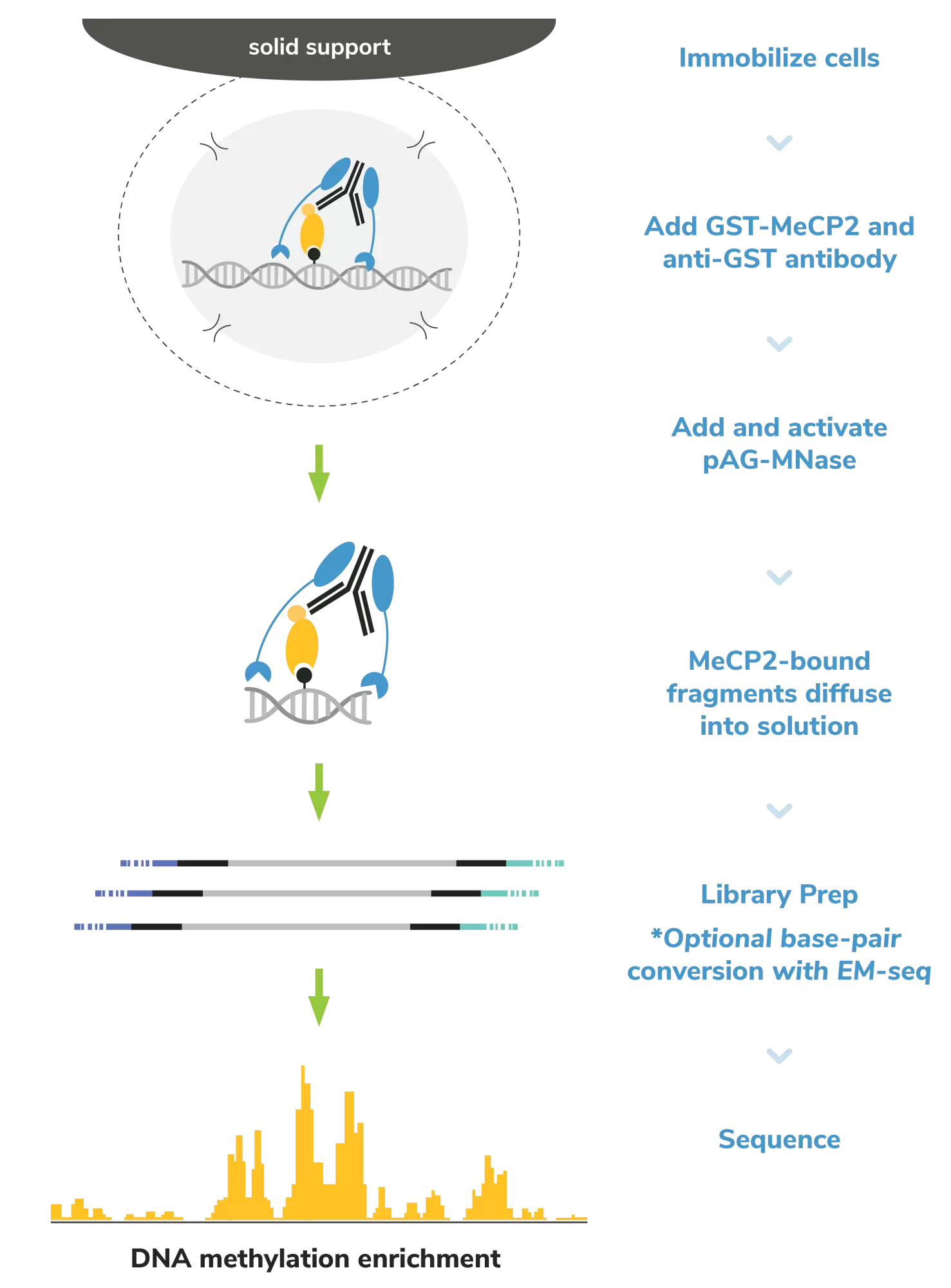
Figure 1: meCUT&RUN methods

