CUTANA™ ChiC / CUT&RUN Assays
Efficient, ultra-sensitive chromatin mapping
CUTANA™ ChIC / CUT&RUN is a high-resolution, cost-effective epigenomic mapping assay that enables sensitive profiling of histone modifications and chromatin-associated proteins using minimal input material.
Compared to ChIP-seq, CUT&RUN offers clear advantages for epigenomic profiling:
- Diverse target profiling : Histone PTMs and chromatin-interacting proteins (including remodelers)
- Reduced cell input : Compatible with as few as 5,000 cells
- Fresh, frozen, and cross-linked cells or nuclei
- Low background: Fewer required sequencing reads per sample (3-5 million)
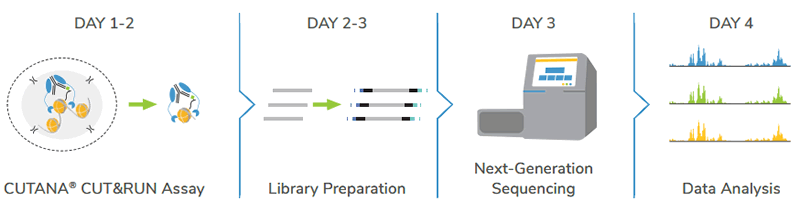
- Cost-effective: Reliable, robust, streamlined workflow (from cells to data in < 4 days)
Go from cells to sequencing using CUTANA™ CUT&RUN or meCUT&RUN and Library Prep Kits
How it works
The Cleavage Under Targets and Release Using Nuclease (CUT&RUN) method builds upon Chromatin ImmunoCleavage (ChIC) technology. In CUT&RUN, a fusion of protein A, protein G and micrococcal nuclease (pAG-Mnase) is used to selectively cleave antibody-labelled chromatin. This strategy eliminates immunoprecipitation steps, greatly simplifying the assay workflow. Clipped chromatin fragments are isolated from solution and used for NGS. The targeted enrichment of CUT&RUN allows for better signal-to-noise with only 3-5 million sequencing reads, significantly reducing sequencing costs vs ChIP-seq.
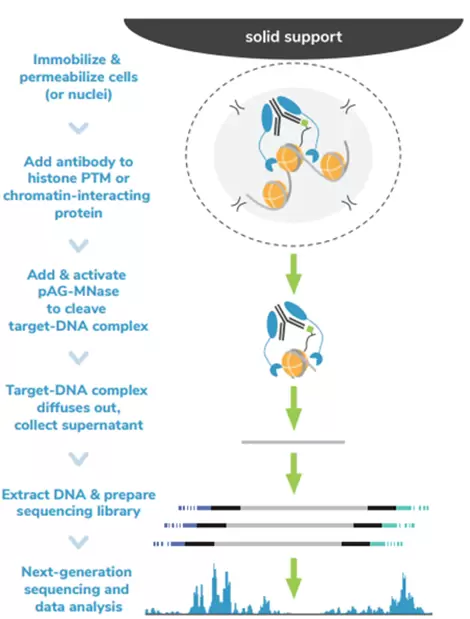
Fig.1. CUTANA™ CUT&RUN Workflow.
CUTANA™ CUT&RUN Platform
The CUTANA™ CUT&RUN platform provides everything you need to get started, including user-friendly kits and protocols, validated antibodies, and more.
CUT&RUN Kits
CUTANA™ CUT&RUN Kit or CUTANA™ meCUT&RUN Kit for DNA Methylation Sequencing
- Includes all the reagents and protocols necessary to perform CUT&RUN assays from cells to DNA
- Compatible with fresh, frozen, or cross-linked cells or nuclei
Library Prep Kits
- High-fidelity library generation for Illumina sequencing
- Cells-to-sequencing solution for chromatin mapping experiments when paired with CUTANA™ CUT&RUN Kit
Antibodies
- Available for various chromatin targets including histone PTMs, transcription factors, and chromatin remodelers
- Lot-validated for robust performance in CUTANA™ CUT&RUN assays
- 4 antibodies available for both CUT&RUN and CUT&Tag assays:
(H3K4me1 Antibody, H3K4me3 Antibody, H3K27me3 Antibody, H3K27ac Antibody)
Reagents
CUTANA™ Reagents and Consumables
- Design and execute your CUT&RUN experiment
- Individual components include ConA beads, pAG-MNase, E. coli spike-in DNA, a DNA purification kit, and many more
Please note: When profiling histone PTMs using fewer than 5,000 cells, we recommend to use CUTANA™ CUT&Tag Assays.
Go from cells to data in < 4 days
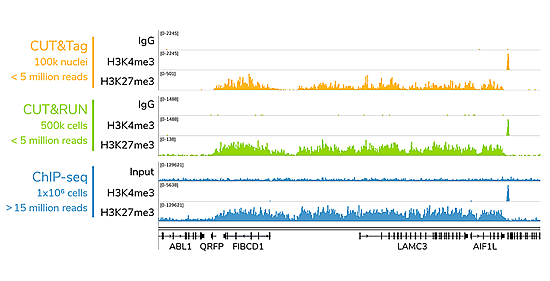
CUT&RUN has superior signal : noise
> 10-fold reduced seq depth compared to ChIP-seq
CUTANA™ assays generate high-quality epigenomic profiles with a fraction of the cells and sequencing depth needed for ChIP-seq. A representative 300 kb region is shown for CUT&Tag (yellow), CUT&RUN (green), and ChIP-seq (blue).