myBaits Custom Methyl-Seq 100-120K
Custom kits for efficient, rapid methylation sequencing of your targets of interest, featuring Daicel Arbor’s optimized probe design and protocol
Product Description
- Unique probe design algorithm for enriching methyl-seq NGS libraries
- Optimized high-performance hyb capture reagents and protocol
- Scaleable to any project size
- Dedicated support from expert scientists
Innovative methyl-seq probe design algorithm was paired with a highly optimized target capture protocol to efficiently enrich target genes and markers of interest from converted methyl-seq NGS libraries. The hybridization capture technique significantly reduces per-sample sequencing costs compared to whole-genome bisulfite sequencing (WGBS) while preserving the essential strand-specific methylation signals.
myBaits Custom Methyl-Seq kits can be paired with either bisulfite or enzymatic-conversion workflows upstream of NGS library preparation, for seamless integration into your existing methyl-seq pipeline. All kits include the latest “v5” generation of myBaits hybridization and wash reagents, pairing best-ever performance with an easy-to-use protocol.
How does myBaits Custom Methyl-Seq work?
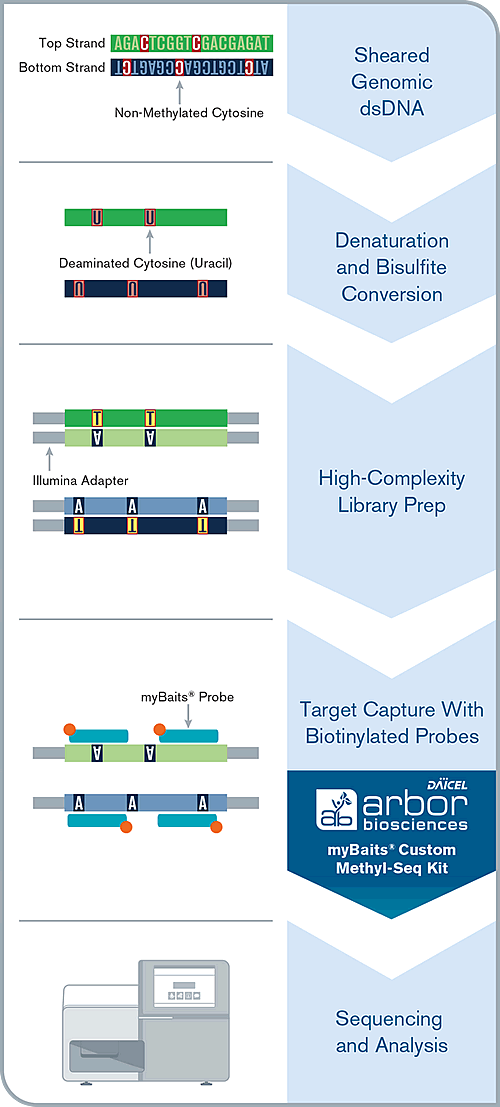
Preparation of Methyl-Seq NGS Library
Genomic dsDNA contains stand-specific patterns of methylated cytosines. In methylation next-generation sequencing, this signal is preserved for NGS by denaturing the two original strands, and separately deaminating any non-methylated cytosines into uracils via bisulfite conversion. This uracils are ultimately transformed into thymines after the original strand is copied by polymerases via amplification. These regenerated dsDNA molecules, with their strand-specific sequences retained, can be converted into NGS libraries, which can be either directly shotgun sequenced, or taken through a hybridization capture procedure.
Hybridization Capture
An NGS library is denatured via heat, and allowed to hybridize to a complex mixture of complementary biotinylated RNA baits over the course of several hours. Adapter-specific blocking oligos prevent random annealing of library molecules at the common adapter sites. After the hybridization is complete, the biotin present on each bait is bound to a streptavidin-coated magnetic bead. Wash steps help remove off-target or poorly-hybridized library molecules. The remaining library molecules that are still bound to their complementary baits are denatured via heat, and amplified using universal library primers. This “enriched” library can now be sequenced.
myBaits Custom Methyl-Seq Performance
Whole genome bisulfite sequencing (WGBS) has become the gold standard in measuring methylation status genome-wide, as it enables methylation signatures to be discovered in a wide range of sample types but its a relatively expensive option. Hybridization capture, which can target thousands to millions of bases of specific genomic sequence in a single reaction, offers an extremely cost-effective solution that enables deeper sequencing and larger studies using a greater number of samples.
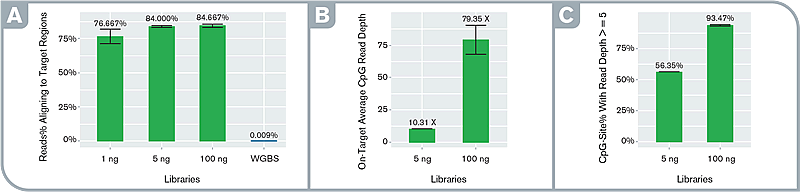
Fig.2 High specificity and orders of magnitude target enrichment. Performance metrics: (A) percentage of on-target reads in enriched and WGBS libraries, (B) average per-site depth of coverage at on-target CpG sites at an even subsample of 950K read-pairs in the enriched libraries, (C) percentage of on-target CpGs with read depth ≥5 at the even subsample in the enriched libraries. Libraries were prepared with the Swift Biosciences Accel-NGS Methyl-Seq Library Kit with varying input amounts and captured with the Daicel Arbor Biosciences myBaits Custom Methyl-Seq system targeting putative promoter regions of 50 cancer-associated genes (total target size ~100Kb).
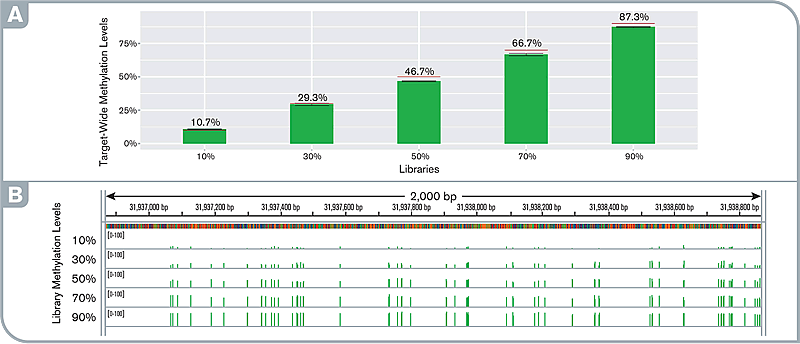
Fig.3 High sensitivity and fidelity of targeted methylation detection at any methylation levels. (A) target-wide methylation levels in pre- and post-capture libraries. Red lines indicate the simulated level. (B) IGV screenshot showing the uniform distribution of methylation levels along the gene STK12 promoter locus observed with the capture libraries at various methylation levels.
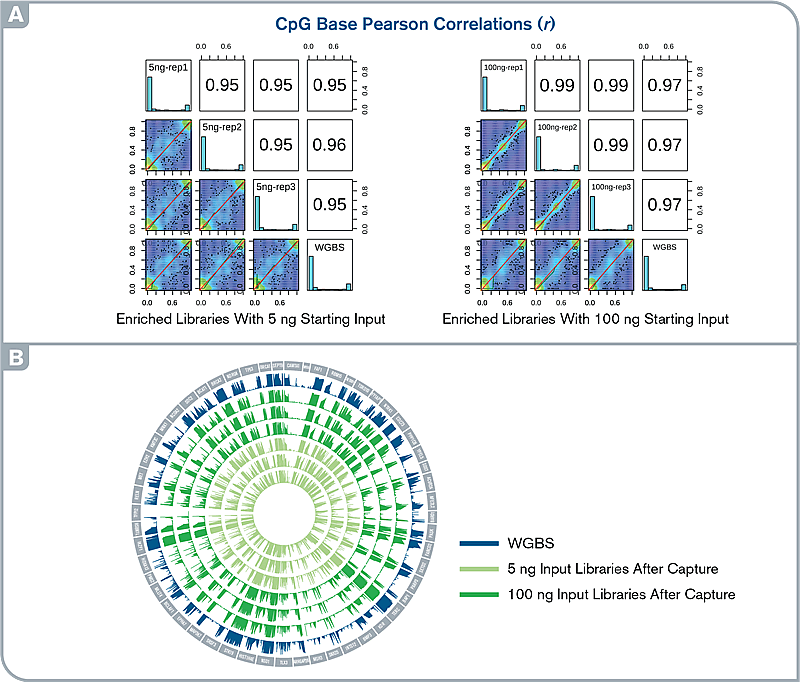
Fig.4 High accuracy and reproducibility of targeted methylation capture. (A) Histograms and scatter plots show the comparison of CpG methylation levels across the target regions between WGBS and capture libraries and among replicates. (B) Circos plot illustrating methylation patterns for the entire target regions in capture and WGBS libraries.
- Catalog Number
300648M.V5-ARB - Supplier
Arbor Biosciences - Size
- Shipping
RT
- Price
- Please inquire

