DriverMap Adaptive Immune Receptor (AIR) Profiling
Get comprehensive and sensitive TCR-seq and BCR-seq profiling results from Bulk or Single Cells
Cellecta's DriverMap™ Adaptive Immune Receptor (AIR) Profiling Assay is the only assay technology on the market for bulk and single-cell T-cell receptor (TCR) and B-cell receptor (BCR) using multiplex RT-PCR and next-generation sequencing (NGS) protocols without the need for any additional specialized equipment.
Cellecta offers bulk DriverMap AIR assays to profile human or mouse RNA, DNA, or both, looking at either full-length receptor regions or only the CDR3 region of all TCR chains--TRA, TRB, TRG, and TRD--and BCR chains--IGH, IGK, and IGL. Additionally, the simultaneous profiling of DNA and RNA from the same sample enables the identification of antigen-activated clonotypes to provide even greater insight into the immune response.
The DriverMap single-cell AIR (scAIR) assays use primers designed to enable full-length receptor sequencing to obtain paired-chain information from individually flow-sorted or diluted T or B cells. This easy-to-use, 96-well plate-based RT-PCR workflow is available for human, mouse, or hybrid human-mouse samples.
Questions?
Connect with us today to get started on your DriverMap AIR Profiling project.
How is DriverMap AIR different from other Adaptive Immune Receptor Repertoire (AIRR) Assays?
- Gene-specific primers are used for DriverMap™ Multiplex PCR technology which significantly reduce the level of non-specific binding and primer-dimer amplification products, and are designed to target only TCR/BCR isoforms.
- Validator barcodes (VBCs) facilitate accurate quantitation of the copy number of cDNA or DNA molecules in amplification steps, as well as detection of low abundance clonotypes and correction of amplification biases and sequencing errors.
- Dual-index amplicon labeling strategy minimizes index hopping during NGS allowing for comprehensive readouts.
- Full profiles of the antigen-recognition CDR3 region enable assessment of CDR3 length distribution, V(D)J segment usage, isotype composition for BCRs, somatic mutations, and similar characteristics with immune receptor profiling software such as MiXCR (MiLabs).
- The Single-cell AIR assay workflow and analysis are economical, with costs starting at 10 cent per cell, not requiring use of a microfluidics platform, complex barcoding protocols, or expensive reagents.
How It Works
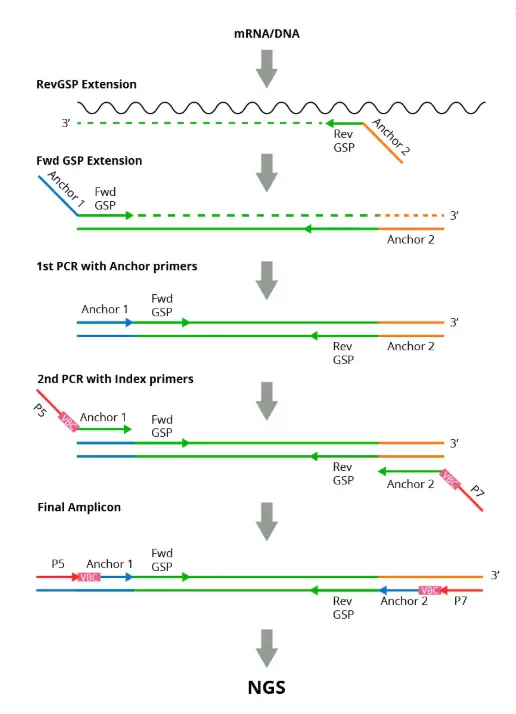
Bulk Assay: Start from RNA or DNA from a variety of immune sample types such as whole blood, PBMCs, cancer biopsies, tissue samples, FFPE or dried blood microsamples.
Single-cell Assay: Start by introducing flow-sorted individual T or B cells or diluted (pipetted) T or B cells into a 96-well plate.
Applications
BCR Sequencing
- Identify broadly neutralizing antibodies (BNAbs) and map Ig-seq datasets to known antibody structures for antibody and vaccine development
- Track B-cell migration and development patterns
- Find clinical markers of autoimmune diseases such as multiple sclerosis, rheumatoid arthritis and cancers (e.g. B-cell lymphoma)
- Contrast naïve and antigenically challenged datasets for understanding antibody maturation
TCR Sequencing
- Track T-cell clonality and diversity for mechanism of action of immune checkpoint inhibitors for immunotherapies
- Assess TCR overlap between repertoires to define spatial and temporal heterogeneity of the anti-tumoral immune response
- Analyze TCR sequence and structure to annotate antigenic specificity for developing personalized cellular immunotherapies
Single-cell TCR or BCR sequencing (scTCR-Seq or scBCR-Seq)
- Characterizing T-cell or B-cell chain pairs and immunophenotypes will aid in the discovery of neo-antigen biomarkers and novel epitopes
- Identifying antigen-specific clonotypesand their functional profiles can guide the design and evaluation of vaccines and immunotherapies, i.e., checkpoint inhibitors and CAR-T cells
- High-throughput screening will be useful in the development of humanized antibodies and for TCR validation studies
Spike-In Controls
To ensure consistent and reproducible T-cell Receptor (TCR) and B-cell Receptor (BCR) clonotype profiling Cellecta designed the DriverMap Adaptive Immune Receptor (AIR) RNA Spike-in Controls. A collection of synthetic mRNA constructs is offered that may be used as universal controls for commercial and home-brewed AIR sequencing assays based on multiplex RT-PCR or 5’-RACE (SMART) PCR techniques.
DriverMap Adaptive Immune Receptor (AIR) TCR-BCR Profiling Services
You would like to profile your immune samples and save precious time? Provide us with information on your samples and your study goals, and we will get back to you to discuss your project.
Get More Info
- Technology Guide
- Improved Adaptive Immune Receptor Repertoire Profiling for Biomarker Discovery (Video, 23:08)
Any Questions Left?
Contact us for assistance to get started on your DriverMap AIR Profiling project.