Whole Genome Bisulfite Sequencing (WGBS) Service
Product Description
Whole Genome Bisulfite Sequencing (WGBS) is a bisulfite sequencing method to detect in-depth DNA methylation across the entire genome, including methylation at CpG sites and less common non-CpG site such as CNG.
WGBS is the gold standard for bisulfite based DNA methylation studies as the entire genome is sequenced with base-level detail into where every methylated cytosine is located.
Features
End-to-End Package
Includes everything from sample preparation to data analysis
Bioinformatics
FASTQ raw data, FASTQC quality control insights, read mapping, methylation calling and differential methylation analysis
Sequencing & Library Preparation Guarantee
Library QC by Bioanalyzer, KAPA quantification and Q30 sequencing score > 75%
High Sequencing Depth
Starting at 20X sequencing depth on PE150 lanes, assuming 1 human library per lane
Post-Completion Technical Support
Complimentary high-quality support to assist with data analysis at no additional charge
Please contact us to discuss your project and get a quote.
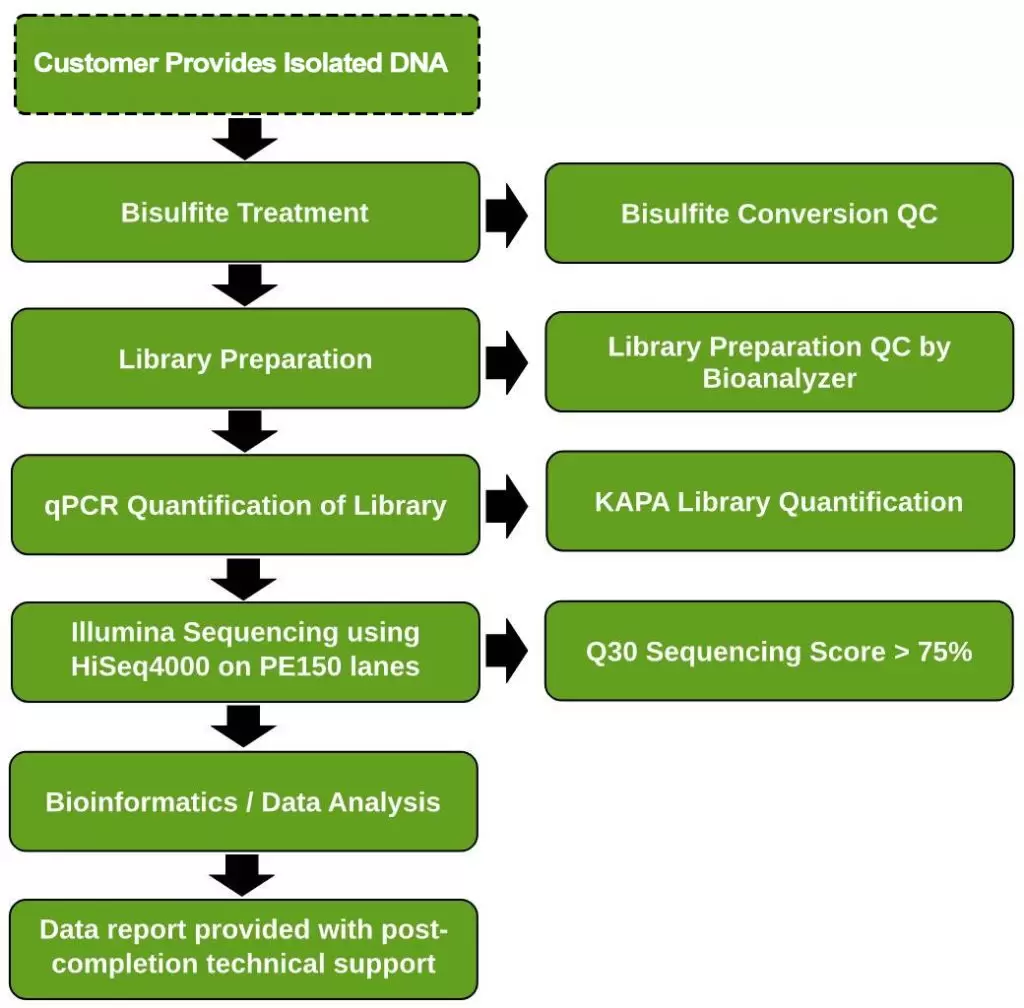
Workflow
Supporting Data
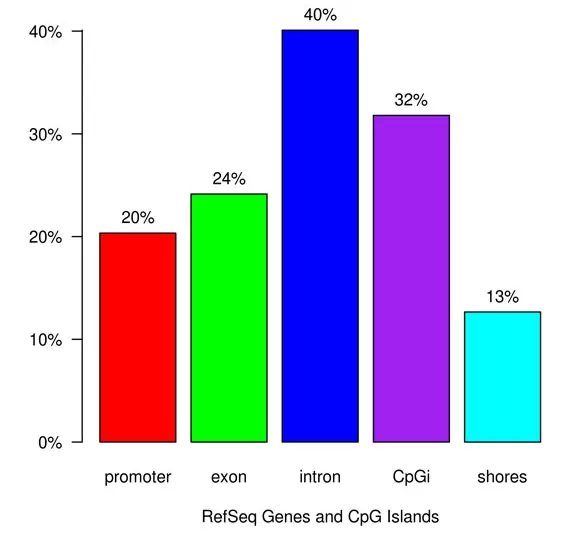
Fig.1. Differential Methylation Annotation: CpG - base. Differential methylation annotation provides a bar chart of differentially methylated sites overlapping with genomic features (promoters, exons, introns, CpG islands, and CpG shores
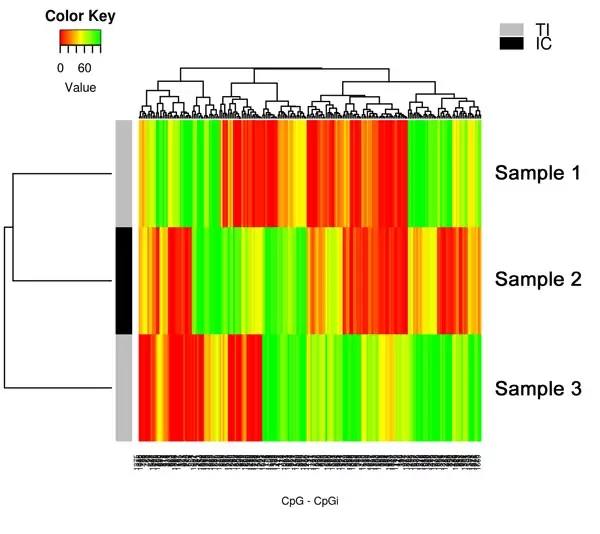
Fig.2. Methylation Heat Map. The Methylation Heatmap offers statistical grouping and visualization of samples based on DNA methylation levels of CpG sites. Green = 100% methylated; Red = 0% methylated.
- Catalog Number
S-1CMS-EP - Supplier
EpigenTek - Size
- Shipping
RT
- Price
- Please inquire

