CM13 Interference-Resistant Helper Phage
Product Description
CM13 is a helper phage especially engineered for phage display. CM13 is a derivative of M13KO7 (1) containing the interference-resistant ir3B A->G mutation (2) at position 8418 of M13KO7. CM13-infected cells often produce more virions than M13KO7 while giving similar levels of display on the minor coat protein III. This helper phage makes preparation of virions particularly efficient at small scale, thus facilitating the overall screening process. This preparation contains enough helper phage to superinfect up to 1 L (1000 ml) of bacterial culture.
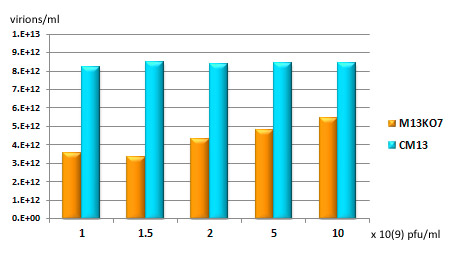
Figure 1. Production of phagemid virions by CM13. pADL-23/HyHEL-10 scFv containing TG1 cells were grown to 1 O.D. (turbidity taken at 600 nm) and transduced with varied amounts of helper phage; after o/n incubation, phagemid virions were prepared by PEG precipitation and quantified by U.V. spectrophotometry; results are expressed in virions per ml of bacterial culture. In this experiment, maximum transduction is reached for CM13 at 1 x 10(9) pfu/ml while larger amounts are needed for M13KO7; CM13 always produced more phagemid virions.
Source
CM13 virions were isolated from the supernatant of infected E. coli TG1 cells and purified by PEG precipitation.
Specifications
- Composition: 50% glycerol TBS buffered
- Concentration: 2 x1012 pfu/ml; 1 x1013 virions/ml; infectivity ~20%
- Storage temperature: -20°C
- Product size: 1 ml.
Quality Control & Certification of Analysis
- Virion concentration: Phage DNA concentrations are determined by UV spectrophotometry and virion concentrations calculated based on the length of CM13 genome.
- Phage titer: CM13 preparations are serially diluted and mixed with melted LB top agar and TG1 cells. The mixtures are poured over LB bottom agar plates and incubated overnight at 37°C; the morning after plaques are counted. Controls include plates without virions and have no visible plaques.
- Infectivity: Infectivities are counted as the ratio between phage titer and virion concentration. Infectivities between 15 and 20% are expected.
- Gene II ORF sequence: ORF DNA is amplified by PCR and sequenced.
- Absence of defective interfering (DI) particles: Integrity of phage particle ssDNA is verified by electrophoresis.
Citations
-
Bordeau B.M., Abuqayyas L., Nguyen T.D., Chen P., Balthasar J.P. (2022). Development and Evaluation of Competitive Inhibitors of Trastuzumab-HER2 Binding to Bypass the Binding-Site Barrier. Front Pharmacol., 13:837744.
-
Stefan M.A., Light Y.K., Schwedler J.L., McIlroy P.R., Courtney C.M., Saada E.A., Thatcher C.E., Phillips A.M., Bourguet F.A., Mageeney C.M., McCloy S.A., Collette N.M., Negrete O.A., Schoeniger J.S., Weilhammer D.R., Harmon B. (2021). Development of potent and effective synthetic SARS-CoV-2 neutralizing nanobodies. MAbs. 13(1):1958663.
-
Wu Y., Chen Z., Sigworth F.J., Canessa C.M. (2021). Structure and analysis of nanobody binding to the human ASIC1a ion channel. Elife, 10:e67115.
-
Gomez-Castillo L., Watanabe K., Jiang H., Kang S., Gu L. (2020). Creating Highly Specific Chemically Induced Protein Dimerization Systems by Stepwise Phage Selection of a Combinatorial Single-Domain Antibody Library. J Vis Exp. , 14;(155):10.3791/60738.
-
Oller-Salvia B., Chin J.W. (2019). Efficient Phage Display with Multiple Distinct Non-Canonical Amino Acids Using Orthogonal Ribosome-Mediated Genetic Code Expansion. Angew Chem Int, 58(32):10844-10848.
-
Tharp J.M., Trae Hampton J., Reed C.A., Ehnbom A., Chen P.C., Morse J.S., Kurra Y., Pérez L.M., Xu S. & Liu W.r. (2020). An amber obligate active site-directed ligand evolution technique for phage display. Nature Comm., 11(1), 1392.
-
He L., Lin X., de Val N., Saye-Francisco K.L., Mann C.J., Augst R., Morris C.D., Azadnia P., Zhou B., Sok D., Ozorowski G., Ward A.B., Burton D.R., Zhu J. (2017). Hidden Lineage Complexity of Glycan-Dependent HIV-1 Broadly Neutralizing Antibodies Uncovered by Digital Panning and Native-Like gp140 Trimer. Front Immunol., 8:1025.

