EpiNext CUT&LUNCH Assay Kit
For ultra-fast, highly specific enrichment of target protein-DNA complexes
Product Description
The EpiNext™ CUT&LUNCH (Cleavage Under Target and Liberate Unique Nucleic Complex Homogeneously) Assay Kit provides a rapid and efficient method for studying in vivo DNA-protein interactions. It integrates all the benefits of ChIP and CUT&RUN while effectively addressing their limitations. The CUT&LUNCH assay has the following advantages and features:
- Rapid Protocol: Complete your assay in less than 2 hours
- Low Non-specific Background: Only 3-5% non-specific DNA binding
- High Specificity & Resolution: DNA is selectively cleaved at both ends of target protein-enriched regions
- Cost-effective: Nearly half the price per reaction compared to traditional CUT&RUN assays.
- No Special Software: Use existing, time-tested ChIP-seq bioinformatics pipelines for NGS data analysis
Background Information
Enrichment of histone or transcription factor (TF)-complexed DNA in vivo, followed by qPCR and/or next-generation sequencing, offers an advantageous tool for studying genome-wide protein-DNA interactions. The major method commonly used to achieve this goal is chromatin immunoprecipitation followed by sequencing (ChIP-seq). This method includes highly specific ChIP-exo and ChIP-nexus. These two assays provide high-resolution mapping. However, the major limitation of these assays is that they need a large amount of input material, cells, or tissue to produce a strong enough signal over background noise. CUT&RUN (Cleavage Under Target & Release Under Nuclease) was developed for mapping protein-DNA interaction with limited biological materials, which requires much less sample amount and also significantly improved mapping resolution. However, a considerable drawback of this assay is non-specific cleavage by antibody un-coupled pAG-MNase, significantly limiting the specificity of the CUT&RUN use for most transcription factors in different species and cell/tissue types. In addition, the original CUT&RUN has complicated steps and needs to optimize assay conditions, which is still time-consuming.
Principle & Procedure
This kit contains all the necessary reagents required for enriching a protein (histone or strong binding transcription factor)-specific DNA complex to profile interactions between proteins and DNA from various cell samples via qPCR or NGS. In this assay, cells are permeabilized and exposed to the ChIP-grade antibody of interest. With the use of a unique nucleic acid cleavage enzyme mix, DNA sequences at both ends of the target chromatin regions are cleaved/removed. The liberated non-specific protein-DNA complexes are eliminated, and only antibody-bound complexes will be selectively recovered. The DNA fragments in the captured protein/DNA complex are purified and can be directly used for gene-specific qPCR or DNA library construction to profile interactions between proteins and DNA.
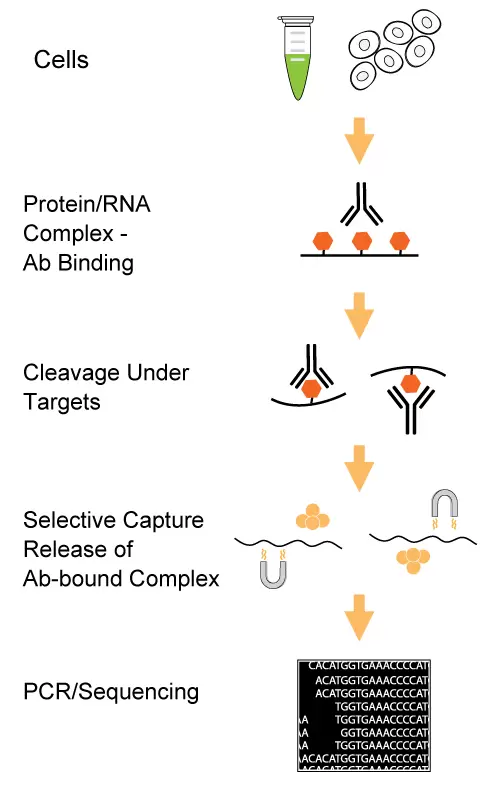
Starting Materials
Starting materials can include various mammalian cell samples such as culture cells from a flask or plate, primary cells, or rare cell populations isolated from blood, body fluid, fresh/frozen tissues, and specific cells sorted from entire cell populations and embryonic cells, etc. The amount of cells can be 2 x 103 to 5 x 105 cells per reaction. For optimal preparation, the cell input amount should be 2 x 105, although the results for modified histones can be obtained with as few as 500 cells.
Performance Data
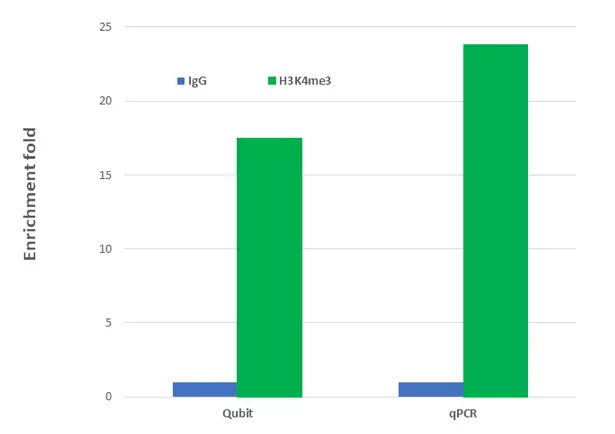
Figure 1: Target protein/DNA enrichment using the EpiNext™ CUT&LUNCH Assay Kit: Histone/DNA complex was captured by the positive control antibody (H3K4me3) from 200,000 MCF-7 cells. Non-immune IgG was used as the negative control. The enriched DNA was purified and fluorescently quantified by Qubit or analyzed by qPCR with targeting to the GAPDH promoter for enrichment fold comparison.
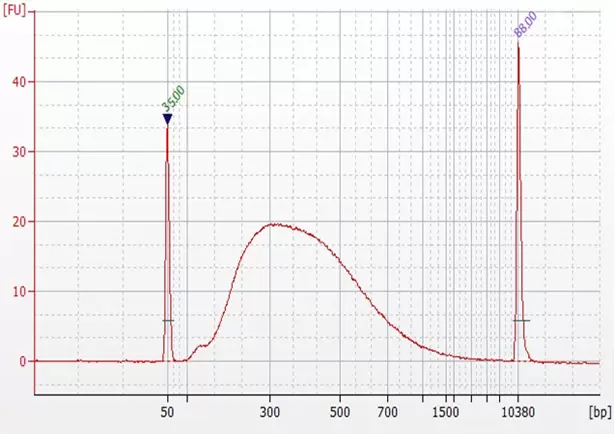
Figure 2: Size distribution of library fragments. Using the EpiNext™ CUT&LUNCH Assay Kit, histone-DNA complex was captured by the positive control antibody (H3K4me3) from 200,000 MCF-7 cells and used for DNA library preparation. The peak around 320 bps reflects the insert size of nucleosome DNA fragments (170 bps).
- Catalog Number
P-2035-24-EP - Supplier
EpigenTek - Size
- Shipping
Blue Ice
You save 25 %
569,00 €
