EpiNext DNA Library Preparation Kit (Illumina)
The kit is suitable for preparing a DNA library for next generation sequencing applications using an Illumina sequencer, which includes genomic DNA-seq, ChIP-seq, MeDIP/hMeDIP-seq, bisulfite-seq, and targeted re-sequencing
Product Description
The EpiNext™ DNA Library Preparation Kit (Illumina) is a complete set of optimized reagents to carry out a successful DNA library preparation. The optimized protocol and components of the kit allow both non-barcoded (singleplexed) and barcoded (multiplexed) DNA libraries to be constructed quickly with reduced bias.
- Fast and streamlined procedure - The procedure from fragmented DNA to size selection is less than 2 h 30 min. Only one clean-up between each step, thereby saving time and preventing handling errors, as well as loss of valuable samples. Gel-free size selection further reduces the preparation time.
- Highly Convenient - The kit contains all required components for each step of DNA library preparation, which are sufficient for end repair, dA tailing, ligation, clean-up, size selection and library amplification, thereby allowing the library preparation to be streamlined with the most reliable and consistent results.
- Minimized bias - Ultra HiFi amplification and an optional PCR-free step enable the user to achieve reproducibly high yields of DNA library with minimal sequence bias and low error rates.
- Flexibility - Can be used for both non-barcoded (singleplexed) and barcoded (multiplexed) DNA library preparation. Uses various dsDNA including fragmented dsDNA isolated from various tissue or cell samples, dsDNA enriched from ChIP reactions, MeDIP/hMeDIP reactions, or exon capture. Broad range of input DNA from 10 ng to 1 µg. PCR-free library preparation can be performed with use of 500 ng or more input DNA.
Background Information
DNA library preparation is a critical step for next generation sequencing (NGS). To generate accurate sequencing data for NGS, the prepared library DNA should be sufficient in yield and of high quality. Also, as NGS technology is continuously improving, DNA library preparation is required to be optimized accordingly.
Most of the currently used methods are unfortunately time-consuming, expensive, and inconvenient. Some of the methods are relatively quick by combining end repair and dA tailing or even ligation in one-step, but have been shown to generate significant G tailing or form concatmers at the ligation step or have high insertion bias. These side reactions eventually result in the prepared DNA library being less efficient and inaccurate.
An ideal DNA library preparation method should be balanced in speed, convenience, small sample-suitability, cost-effectiveness, and accuracy. To address this issue, Epigentek offers the EpiNext DNA Library Preparation Kit.
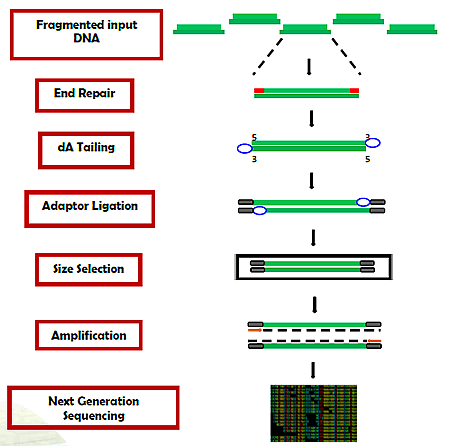
Fig. 1. Workflow of the EpiNext™ DNA Library Preparation Kit (Illumina).
Principle & Procedure
This kit incluldes all reagents required at each step to carry out a successful DNA library preparation. In the library preparation, DNA is first fragmented to the appropriate size (about 300 bp peak size).
The end repair of the DNA fragments is performed and an A-overhang is added at the 3'-end of each strand. Adaptors are then ligated to both ends of the end repaired/dA tailed DNA fragments for amplification and sequencing.
Fragments are then size selected and purified with MQ beads, which allows for quick and precise size selection of DNA. Size-selected DNA fragments are then amplified with a high-fidelity PCR Mix which ensures maximum yields from minimum amounts of starting material and provides highly accurate amplification of library DNA with low error rates and minimum bias.
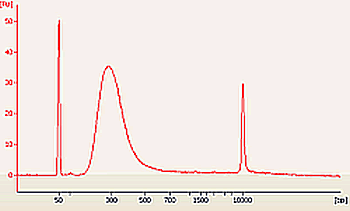
Fig. 2. Size distribution of library fragments: human placenta DNA was sheared to 210 bps peak size and 20 ng of sheared DNA was used for DNA library preparation using the EpiNext™ DNA Library Preparation Kit (Illumina).
Starting Materials
Starting materials can include fragmented dsDNA isolated from various tissue or cell samples, dsDNA enriched from ChIP reactions, MeDIP/hMeDIP reaction, or exon capture. DNA should be relatively free of RNA since large fractions of RNA will impair end repair and dA tailing, resulting in reduced ligation capabilities.
Input amount of DNA can be from 5 ng to 1 µg. For optimal preparation, the input amount should be 100 ng to 200 ng. For amplification-free, 500 ng or more is needed.
- Catalog Number
P-1051-12-EP - Supplier
EpigenTek - Size
- Shipping
Blue Ice

