Small RNA Library and Sequencing Service for total RNA
Product Description
RealSeq®-AC Technology for small RNA library preparation from total RNA or from tissue/cell cultures. RealSeq®-AC is optimized for inputs between 1 ng and 1000 ng of total RNA from tissue or cells.
Service Features
- For total RNA samples and biological samples (tissue and cells)
- Superior quality sequencing libraries
- Sequencing on an Illumina platform (MiniSeq or Next Seq)
- Avoids underdetection of many miRNAs
- Service includes:
1) Small RNA library prep from total RNA (or from tissue/cell cultures), using RealSeq-AC
2) Next-generation sequencing of small RNAs (10 Million reads/sample)
Supporting Data
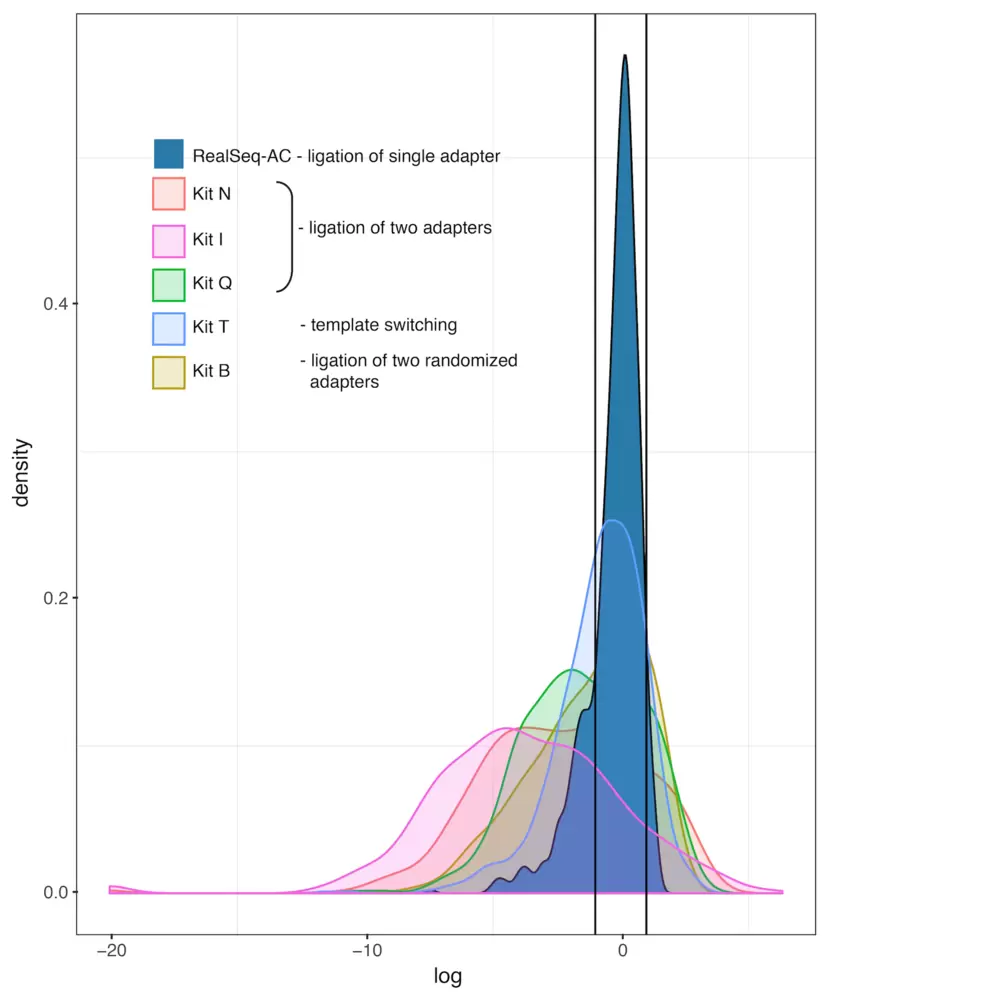
Fig.1. One pmole of miRXPlore Universal Reference pool (Miltenyi Biotec) was used to compare incorporation bias in six different commercially available library preparation kits. Purified libraries were sequenced on the Illumina MiSeq platform. Trimmed sequencing reads were aligned to a custom miRNA reference. Reads mapping to miRNAs were counted and fold-deviations from the equimolar input were calculated and plotted as log2 values. Measurements of miRNA levels within a factor of two of the expected values (between vertical lines) are considered unbiased (Fuchs et al, 2015). The method of adapter attachment to the miRNAs is noted in the legend.
RealSeq®-AC is highly efficient, detecting more miRNAs in total RNA samples
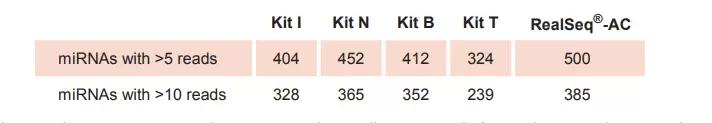
Table 1. RealSeq®-AC detects more miRNAs with over 5 or 10 reads per million, respectively, from total RNA samples compared to other kits for miRNA sequencing library preparation. RealSeq®-AC is optimized for inputs between 1 ng to 100 ng of total RNA. Lower numbers of PCR cycles are reducing PCR-induced issue.
- Catalog Number
500-totalRNA-SOM - Supplier
RealSeq Biosciences - Size
- Shipping
RT
- Price
- Please inquire

