
Gene qPCR Arrays
High-Throughput Gene Expression Profiling
The ExProfile™ Gene qPCR Arrays are designed for profiling the expression of pre-defined sets of coding genes in various tissues or cells. They profile the expression of cancer-related, pathway-related or disease-related genes as well as of functional gene groups, which are carefully chosen for their close respective correlation based on a thorough literature search of peer-reviewed publications. The resulting differential expression of profiled genes helps researchers to identify those that are biologically significant and relevant to their research. In each 96-well plate, there are up to 84 pairs of qPCR primers and 12 wells of controls which are used to monitor the efficiency of the entire experimental process – from reverse transcription to qPCR reaction.
Each pair of primers used in the qPCR arrays has been experimentally validated to yield a single dissociation curve peak and to generate a single amplification of the correct size for the targeted mRNA. A cDNA pool, containing reverse transcript products from total RNA of 10 different tissues, was used as the qPCR validation template.
Universal real-time PCR conditions were developed for easy profiling and analysis of gene expression in a high-throughput fashion. The All-in-One™ First-Strand cDNA Synthesis Kit and qPCR Mix Kit are the recommended and supported RT-PCR reagents for use with the ExProfile™ Gene qPCR arrays. These reagents have been optimized to produce high sensitivity, efficiency, and specificity.
- Profile cancer genes, pathway genes, disease genes or gene groups
- Validated primers designed with proprietary algorithm
- Detect as low as 4 copies of mRNA
- Simultaneously detect mRNAs at different expression levels
- High reproducibility (R2 > 0.99) for inter-array and intra-array replicates
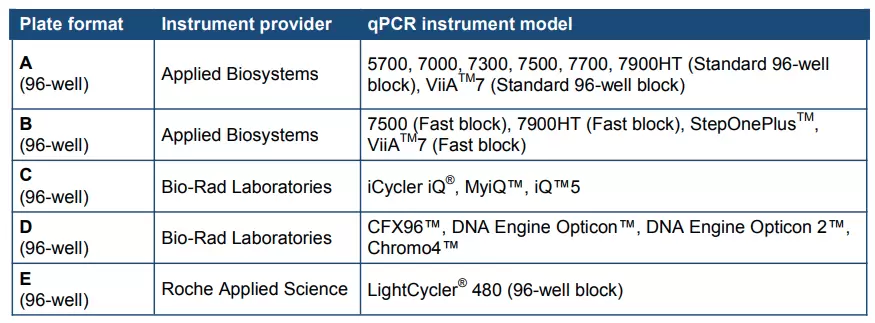
Array Format
Five qPCR array formats (A, B, C, D, and E) suitable for use with the following real-time PCR instruments are provided. Before placing an order, please take care to choose the right kit version which is compatible with your instrument.
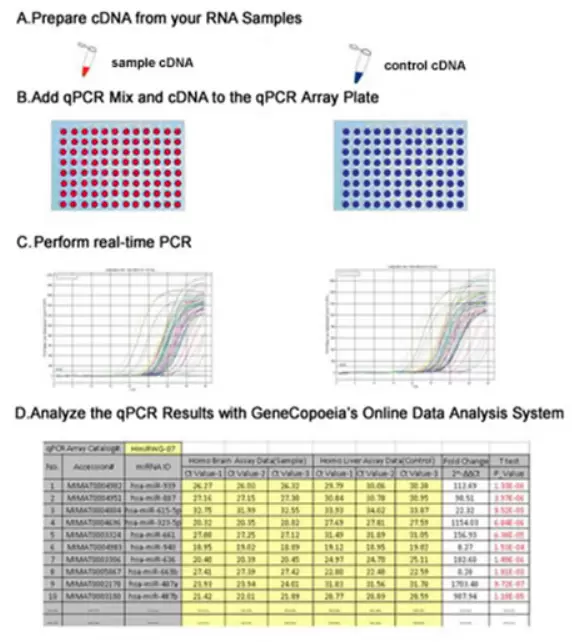
How It Works
Gene qPCR Array experimental workflow
Performance Data
Linear Range and Sensitivity (Spike-in control RNA)
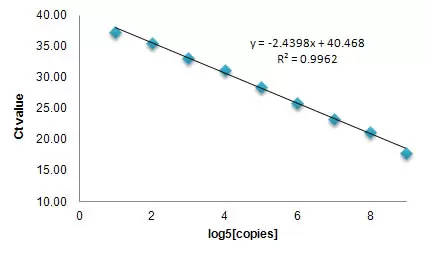
Broad linear range and high sensitivity. Mouse total RNA with serially diluted Spike-in control RNA were reverse-transcribed using All-in-One first strand cDNA synthesis kit. The reverse-transcribed cDNA samples were detected using All-in-One qPCR mix and spike-in control specific primers deposited in a 96-well plate. The resulting Ct values were plotted against the log5 of the amounts of spike-in control RNA. The data demonstrated a broad linear dynamic range from 4 to 1.6*106 copies of input RNA as well as high sensitivity.
Positive Calls with a Range of total RNA
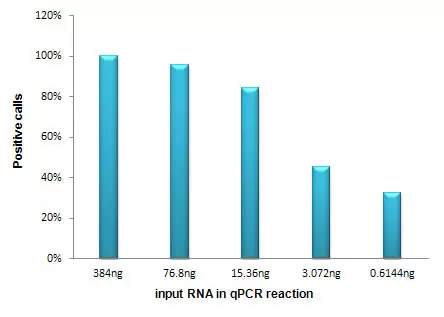
High positive calls with as little as 15.36 ng of total RNA. Different amounts of MCF_7 total RNA (1000, 200, 40, 8, 1.6ng) were analyzed with the Human Breast Cancer Gene qPCR Array.The percentage of positive calls (Ct < 35) is plotted against the input amount of total RNA in each qPCR reaction.
Inter-Array Reproducibility
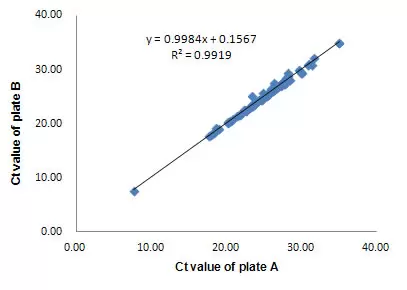
High inter-array reproducibility. Two ExProfile™ qPCR gene array replicates (plate A and B) were analyzed using human total RNA (10-tissue mix) on the Bio-Rad iQ5. The Ct values of the replicate plates were plotted against each other. R2 > 0.99 was observed for high inter-array reproducibility. R2 > 0.99 was also observed for intra-array reproducibility (data not shown).