
myBaits Hybridization Capture Kits
Hybridization Capture using biotinylated RNA Baits
myBaits® target capture kits can greatly enhance the efficiency and cost-effectiveness of NGS research projects for any organism and any project size
The Kits from Daicel Arbor Biosciences provide pools of in-solution biotinylated RNA probes plus reagents for highly-efficient, scalable targeted sequencing on any NGS platform, including Illumina®, Ion Torrent®, PacBio®, and Nanopore®. Daicel Arbor's proprietary oligo synthesis technology delivers high-quality, complex baits at extremely competitive pricing. In addition, we can offer complimentary bait design and project development assistance from a team of expert scientists.
myBaits kits have been successfully used in thousands of research projects from a wide variety of genome types (e.g. animals, plants, viruses and microbes) and DNA sources (e.g. fresh, degraded, and environmental).
Many biological sample types are dominated by DNA from other sources, requiring significant next-generation sequencing (NGS) depths to resolve specific microbial genomes or to fully characterize the variation within microbial communities. Targeted sequencing that excludes background non-target DNA from samples prior to NGS solves this problem and dramatically reduces the overall costs of sequencing and data analysis per sample. Hybridization capture is the most versatile technique for comprehensive, cost-effective targeted NGS of e.g. viruses and bacteria in complex samples.
Key Benefits
- Specificity and sensitivity - myBaits chemistry delivers a high percentage of on-target reads (specificity) without sacrificing the unique target molecule complexity (sensitivity)
- Novel variant discovery - Detect any amenable mutation type, including SNPs, indels, rearrangements, CNVs, and more
- Capture any molecule - Since myBaits probes are completely independent of the enriched library, they can capture molecules of any length
- Flexible workflows meet your needs - myBaits kits are designed to be paired with ours or any other upstream library prep kit and any downstream NGS platform
- Detect rare target molecules - High-sensitivity protocol enables high specificity of rare targets from challenging samples without sacrificing sensitivity
- Preserve relative abundance - Unlike other targeted sequencing techniques, myBaits maintains the relative abundance signals between target types with minimal bias
- Wide range of target types - Daicel Arbor’s flexible oligo synthesis enables focused NGS for any total target size, from a single locus to hundreds of megabases
- Flexible reagents - You can deploy your myBaits in-solution reagents in any manner to fit your project, whether running one or hundreds of captures at once
myBaits Custom Kits
Kits tailored for specific target regions are the most popular choice, allowing enrichment of specific SNPs, exons, genes, and other sequence motifs from genomic or metagenomic samples.
- Target Capture Kits for DNA- and RNA-Seq
- Target Capture Kits for Methyl-Seq
- Custom Whole Genome Enrichment (WGE) Kit
myBaits Expert Kits with Predesigned Panels
Daicel Arbor Biosciences also offers a variety of predesigned kit options for specific research applications, such as mitochondrial DNA sequencing, ultraconserved element sequencing, whole-genome enrichment from metagenomic samples, and human cancer exome research.
- Virus - SARS-CoV-2
- 16S-Hyb
- Wheat Exome
- Mito Panel
- Onconome
- UCE (Ultraconserved Elements)
- Plant - Angiosperms
- Plant - Compositae
- Human Whole Genome Enrichment (WGE)
How does myBaits work?
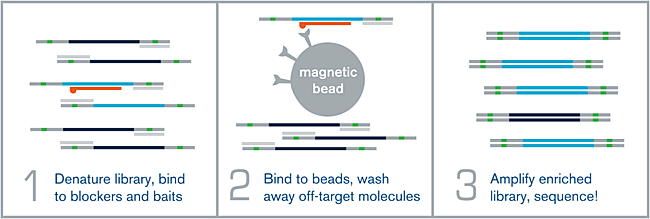
Figure 1.
(1) HYBRIDIZATION – An NGS library is denatured via heat, and allowed to hybridize to a complex mixture of complementary biotinylated RNA baits over the course of several hours. Adapter-specific blocking oligos prevent random annealing of library molecules at the common adapter sites.
(2) WASHING – After the hybridization is complete, the biotin present on each bait is bound to a streptavidin-coated magnetic bead. Wash steps help remove off-target or poorly-hybridized library molecules.
(3) AMPLIFICATION – The remaining library molecules that are still bound to their complementary baits are denatured via heat, and amplified using universal library primers. This “enriched” library can now be sequenced.
Understanding myBaits Custom Performance
Hybridization capture with myBaits Custom is an extremely versatile and flexible technology that has been applied to an enormous variety of applications and taxa, such as plants, animals, humans, bacteria, and viruses.
Hundreds of studies have been published with myBaits over the years. Across the wide variety of applications, myBaits Custom kits frequently achieve high on-target read percentages and/or high unique read complexity for most applications, creating many orders of magnitude savings in sequencing compared to shotgun approaches.
In addition, research scientists are continually innovating novel improvements to the speed, ease of use, and performance of our myBaits kits across a variety of applications.
We can always provide specialized experimental design advice to maximize success with your next NGS targeted sequencing project. Our Sequence Submission Guidelines describe how you need to provide your sequence data for the design of your myBait set.
Typical example of visualized NGS reads
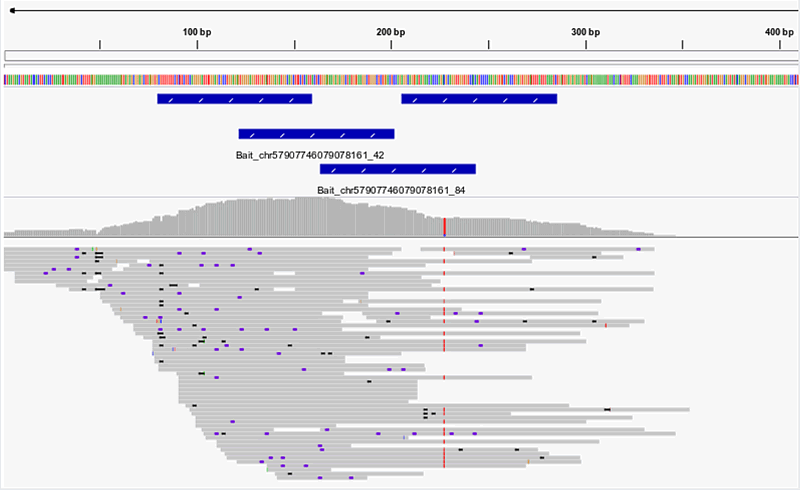
Figure 2. NGS reads from enriched human gDNA library. Single-end NGS reads are shown aligned to the hg38 genomic reference sequence. Positions of the four 80nt myBaits probes in this region are indicated by blue bars. Unique read coverage is highest across the baited region, and tapers off upstream and downstream of the baited region. If sequencing further into the known or unknown flanking regions is desired, simply increase the length of your NGS library molecules (compatible with any myBaits kit).
Selected Citations
- Köndgen, S. et al. (2024) A robust, scalable, and cost-efficient approach to whole genome sequencing of RSV directly from clinical samples. Journal of Clinical Microbiology 0, e01111-23. https://doi.org/10.1128/jcm.01111-23
- Matiasek, K. et al. (2023) Mystery of fatal ‘staggering disease’ unravelled: novel rustrela virus causes severe meningoencephalomyelitis in domestic cats. Nat Commun 14, 624. https://doi.org/10.1038/s41467-023-36204-w
- Ruscher, C. et al. (2023) Ecological and clinical evidence of the establishment of West Nile virus in a large urban area in Europe, Berlin, Germany, 2021 to 2022. Eurosurveillance 28, 2300258. https://doi.org/10.2807/1560-7917.ES.2023.28.48.2300258
- Ulrich, L. et al. (2022) Enhanced fitness of SARS-CoV-2 variant of concern Alpha but not Beta. Nature 602, 307–313. https://doi.org/10.1038/s41586-021-04342-0
- Weiss, S. et al. (2022) Kiwira Virus, a Newfound Hantavirus Discovered in Free-tailed Bats (Molossidae) in East and Central Africa. Viruses 14, 2368. https://doi.org/10.3390/v14112368
- Wertheim, J.O. et al. (2021) Discovery of Novel Herpes Simplexviruses in Wild Gorillas, Bonobos, and Chimpanzees Supports Zoonotic Origin of HSV-2. Molecular Biology and Evolution 38, 2818–2830. https://doi.org/10.1093/molbev/msab072
- Wylezich, C. et al. (2021) Next-generation diagnostics: virus capture facilitates a sensitive viral diagnosis for epizootic and zoonotic pathogens including SARS-CoV-2. Microbiome 9, 51. https://doi.org/10.1186/s40168-020-00973-z
- Binder, F. et al. (2020) Isolation and characterization of new Puumala orthohantavirus strains from Germany. Virus Genes 56, 448–460. https://doi.org/10.1007/s11262-020-01755-3
- Hiltbrunner, M. and Heckel, G. (2020) Assessing Genome-Wide Diversity in European Hantaviruses through Sequence Capture from Natural Host Samples. Viruses 12, 749. https://doi.org/10.3390/v12070749
- Forth, J.H. et al. (2019) A Deep-Sequencing Workflow for the Fast and Efficient Generation of High-Quality African Swine Fever Virus Whole-Genome Sequences. Viruses 11, 846. https://doi.org/10.3390/v11090846